Multi-Scale Context Aggregation for Strawberry Fruit Recognition and Disease Phenotyping
Published in IEEE Access, 2021
Ilyas Talha1, Khan Abbas1, Umraiz Muhammad1, Kim Hyongsuk1,2,*, Jeong Yongchae1,2,*
1Division of Electronic Engineering, Intelligent Robots Research Center, Jeonbuk National University, Jeonju, South Korea
2Division of Electronic and Information Engineering, Jeonbuk National University, Jeonju, South Korea
*Corresponding author: hskim@jbnu.ac.kr
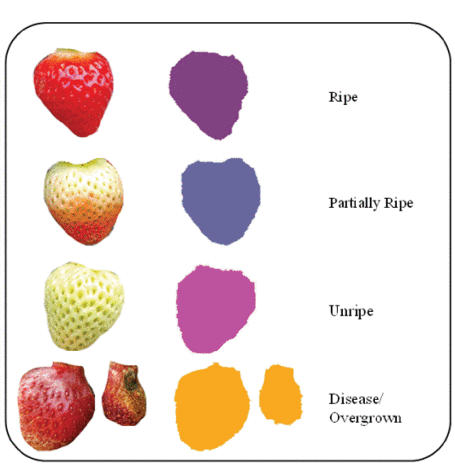
In this paper, a new dataset (i.e., SS1K) is introduced for the segmentation of strawberries into four classes depending upon the ripeness of the fruit (including a background class). The proposed segmentation network named Straw-Net improves the performance of ASHs in unconstrained and natural farming environments.
Abstract
Timely harvesting and disease identification of strawberry fruits is a major concern for commercial level cultivators. Failing to harvest the grown strawberries can result in the fruit rotting which makes their damaged tissues more prone to grey mold pathogens. Immediate removal of the overgrown or diseased strawberries is inevitable to curb the mass spreading of the pathogen. In this paper, we propose a deep learning-based framework to identify three different strawberry fruit classes (unripe, partially ripe and ripe), as well as a class of overgrown or diseased strawberries. We equip the proposed convolutional encoder-decoder network with three different modules. One for adaptively controlling receptive filed size of the network to detect objects of multiple sizes. Second for controlling the flow of salient features (information) to the deeper layers of the network and the other for controlling the architecture’s computational complexity. These modules combined, outperform the previous state-of-the-art semantic segmentation networks on the task of strawberry fruit phenotyping. We also introduce a dataset collected from different farms to evaluate the performance of the network. Quantitative and qualitative results show that notwithstanding heterogeneity in the data and the effect of the real-field variations, our approach produced remarkable results with a 3% increase in mean intersection over union as compared to the other state-of-the-art networks and was able to recognize diseased fruits with a precision of 92.45%.
